GSoC 2024 Final Project Report

Table of contents
- Table of contents
About Fossology
FOSSology is quite an old and mature project. Few years ago, Fossology could only be used in mainly two ways: As a toolkit, the scanners binaries are provided that can be executed as a CLI tool. And as a system, a database and web UI are provided to scan repos, directories, packages etc.
Few years ago, my mentors had an idea of providing the fossology scanners in CI environments.
That led to developing a fossology/fossology:scanner docker image that is capable of running license
checks (using nomos or ojo) and keyword and copyright scans.
Motivation for my project
This CI image was relatively new and needed numerous quality of life improvements:
- The output of the script was not easy to understand as it did not pinpoint exactly where the license had been found by the scanner.
- The script scanned the repos in mainly two modes:
diffmode andrepomode. There was no method for comparing two different tags (or versions) of the project. - For ignoring some licenses from scanning, fossology provides a whitelisting feature but it was difficult to customize it.
- The
keywordscanner scanned for a predefined set of keywords in the repository which needs to be customizable as each project might want to scan for specific keywords. - Most of the projects have some dependencies, which also needed to be scanned. Support for scanning dependencies was crucial for compliance check.
The motivation of my project was to tackle these existing issues and provide a neat and scalable solution for the same.
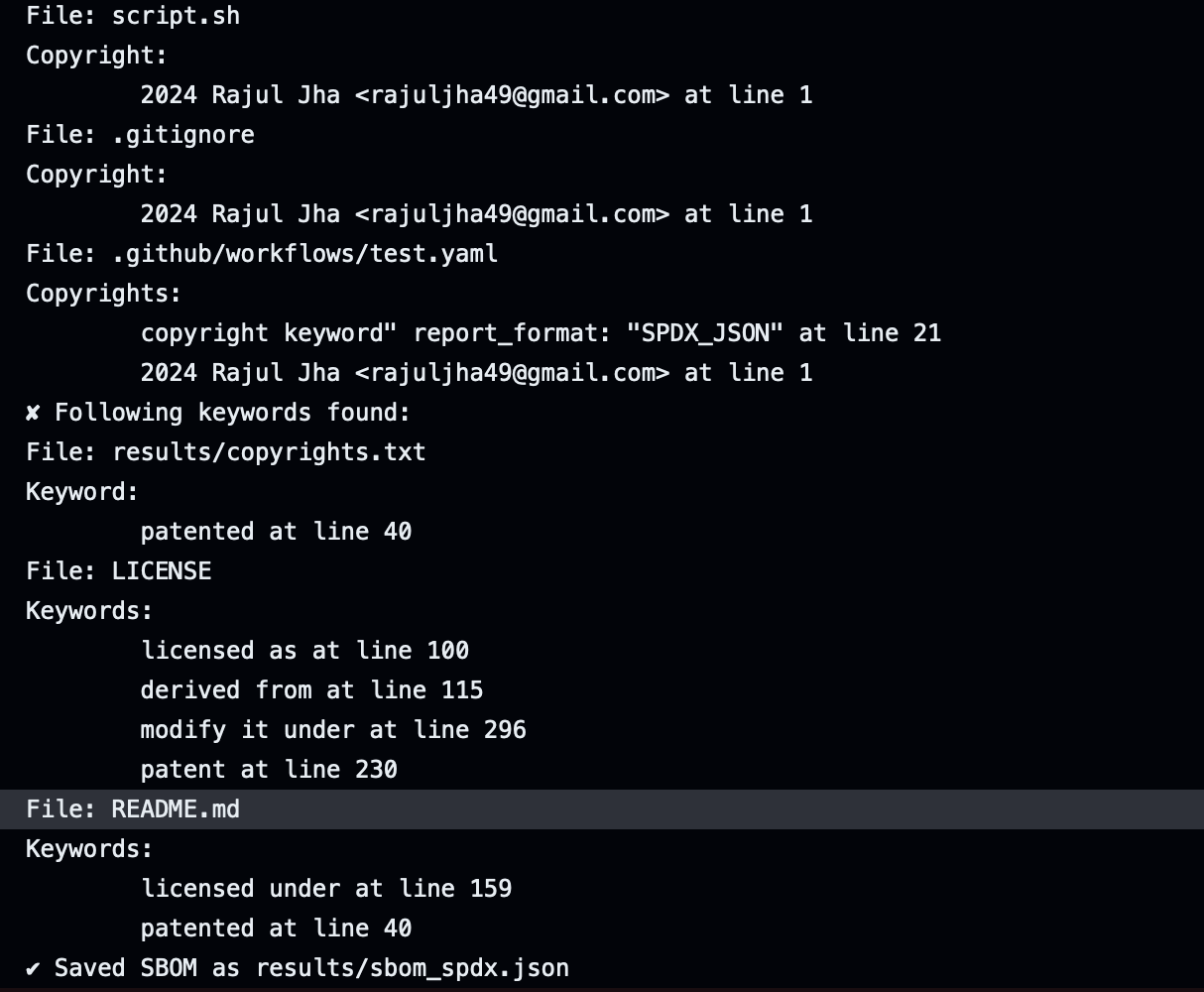
Providing line numbers to the results
To improve the readability of the results and pinpoint the license location, we decided to provide the line number of licenses in the scan results. The first thing to do was to figure of an algorithm for calculating the line numbers of the license. Each scanner stored the start_byte and end_byte information of the results, which could be used for this purpose.
The algorithm can be broken down into two parts based on the scanning modes:
Repo/Directory Mode: It was pretty straightforward as to counting the occurence of line breaks (
\n) in the raw results of the scanners.Diff Mode: For diff mode, we use the git diff’s to fetch the diff and scan on them using respective API’s. This posed a problem. How can we get the corrected line number from the diffs?

The solution was to convert the the line numbers to unified diff format and then get the actual line number from the modified result.
git diff "$@" | gawk ' match($0,"^@@ -([0-9]+),([0-9]+) [+]([0-9]+),([0-9]+) @@",a){ left=a[1] ll=length(a[2]) right=a[3] rl=length(a[4]) } /^(---|\+\+\+|[^-+ ])/{ print;next } { line=substr($0,2) } /^[-]/{ padding = right; gsub(/./, " ", padding); printf "-%"ll"s %"rl"s:%s\n",left++,padding,line; next } /^[+]/{ padding = left; gsub(/./, " ", padding); printf "+%"ll"s %"rl"s:%s\n",padding,right++,line; next } { printf " %"ll"s %"rl"s:%s\n",left++,right++,line } 'I translated the gawk command to a python script and viola. It worked!
The nomos scanner did not have the byte information in the json output. I extended it’s functionality
to add the required fields.
The results are formatted in this way:
Providing custom allowlisting and keywords scanning
Allowlisting
Introduced a new CLI flag --allowlist-path that taken in the path to the custom allowlist.conf file
stored in the repository of where the CI is running. Moreover, it deprecates using the word whitelist
by printing a deprecation warning.
The decision tree looks like this:
.png)
Keyword Scanning
Also introduced the functionality of providing custom set of keywords for scanning. Previously the
keyword scanner used the predefined set of keywords for scanning defined here.
I introduced a new CLI flag --keyword-conf to scan for a set of keywords defined at a particular path.
I also updated the github and gitlab workflow examples in the repository.
Also added some validation checks for the file as the keyword scanner might break if an invalid file is
passed.
Differential Scans
Previously, the scanner image supported two types of scanning modes, repo and diff. I introduced a
new scanning mode, differential, which can scan for any two tagged versions of the repository.
It works with the --tags CLI flag.
For example,
fossologyscanner nomos ojo differential --tags v001 v004
This will scan in differential mode between the v001 and v004 tags of the repository.
It utilizes the Github and Gitlab REST API’s respectively to fetch the diff content between them.
They use the following endpoints to do so:
1. https://api.github.com/repos/<username>/<repo_name>/compare/v001...v004
2. https://gitlab.com/api/v4/projects/:projectid/repository/compare/?from=v001&to=v004
Fossology Github Action
The fossology docker image required a way to easily integrate the fossology scanners in the existing workflows. For making this process easier, I created a composite Fossology Github Action, which integrates scanning with fossology into GH Action workflows.
The user can pass a number of arguments to achieve any custom output to meet their needs.
scan_mode:
description: "Specifies whether to perform diff scans, repo scans, or differential scans.
Leave blank for diff scans."
required: false
default: ""
scanners:
description: "Space-separated list of scanners to invoke."
required: true
default: "nomos ojo copyright keyword"
report_format:
description: "Report format (SPDX_JSON,SPDX_RDF,SPDX_YAML,SPDX_TAG_VALUE) to print the results in."
required: false
default: ""
keyword_conf_file_path:
description: "Path to custom keyword.conf file. (Use only with keyword scanner set to True)"
required: false
default: ""
allowlist_file_path:
description: "Path to allowlist.json file."
required: false
default: ""
from_tag:
description: "Starting tag to scan from. (Use only with differential mode)"
required: false
default: ""
to_tag:
description: "Ending tag to scan to. (Use only with differential mode)"
required: false
default: ""
Example workflows
- Pull request scans
name: License scan on PR
on: [pull_request]
jobs:
compliance_check:
runs-on: ubuntu-latest
name: Perform license scan
steps:
- name: Checkout
uses: actions/checkout@v2
- name: License check
id: compliance
uses: fossology/fossology-action@v1
with:
scan_mode: ''
scanners: 'nomos ojo'
report_format: 'SPDX_JSON'
- Tag scans
name: License scan on tags
on: [tags]
jobs:
compliance_check:
runs-on: ubuntu-latest
name: Perform license scan
steps:
- name: Checkout
uses: actions/checkout@v2
- name: License check
id: compliance
uses: fossology/fossology-action@v1
with:
scan_mode: 'differential'
scanners: 'nomos ojo copyright keyword'
from_tag: 'v003'
to_tag: 'v004'
report_format: 'SPDX_JSON'
Dependencies Scanning
Every major project is shipped with some dependencies or the other. Hence, we need a method to scan these
dependencies inside the CI environment itself. I tried to tackle this issue by providing a functionality
to pass a SBOM file via --sbom-path flag. We take the file, and pass it to a main Parser Object, whose
job is to sort the dependencies based on languages.
Then there are language dependant parsers for files like PythonParser, NPMParser and PHPParser.
They take in each component and extract the download urls for each package from them and pass that to the
Downloader class.
The downloader class takes the download urls and downloads the packages locally
concurrently using ThreadPoolExecutor.
The dependencies can be scanned mainly in two ways:
1. fossologyscanner nomos ojo scan-only-deps --sbom-path <path_to_sbom_file> (for scanning only dependencies.)
2. fossologyscanner nomos ojo repo --sbom-path <path_to_sbom_file> (for scanning repo along with dependencies.)
They can be used in any combination with existing options to fit your use case.
Relevant PR’s
- feat(automation) : Add line numbers to keyword and copyright scanners
- feat(automation) : Add custom keyword.conf file
- feat(differential scans): Add differential scans to CI Scanner
- feat(automation): Add custom allowlist.json
- chore(fossology action): Add fossology action
- feat(nomos): Add start, end, len to nomos JSON output.
- feat(dir_scan): Scan a subdirectory inside CI
- feat(Dependecy Scan): Add Python Dependency Scan
Additional PR’s
- refactor(automation): Upgrade spdx_tools package from 0.8.0a2 to 0.8.2
- hotfix(docker_test): Use docker compose (v2) instead of docker-compose (v1)
Throughout GSoC, I have maintained a work log in the form of weekly progress report that is available at the Fossology GSoC Page
Deliverables
| Tasks | Planned | Completed |
|---|---|---|
| Provide line numbers to scanner output | Yes | ✅ |
| Add allowlisting from custom location | Yes | ✅ |
| Add custom keyword scanning functionality | Yes | ✅ |
| Add differential scans feature | Yes | ✅ |
| Add dependencies scanning for major languages | Yes | ✅ (partially) |
| FOSSology Github Action | No | ✅ |
Known drawbacks
- The copyright scanner results give some unnecessary information along with the copyright findings, which is a known issue of the copyright scanner. This needs to be fixed in coming future.
- The line number algorithm might struggle to find the line numbers if the diff is not properly formatted or is tampered with. It heavily depends on the diff format.
My Learnings
- Git was definitely the skill I improved the most during GSoC :)
- I spent a lot of time working with Python, Docker, and CI/CD tools like GitHub Actions, and I feel way more confident in using them now.
- Gained hands-on experience with SBOM generation, package parsing, and the integration of FOSSology scanners, which broadened my technical expertise.
- I learned some valuable lessons on writing clean, maintainable code. I focused on proper formatting, modular programming, and object-oriented techniques.
- I also got to learn about different packaging methods that are industry standards and why following community norms is so important. It’s the little things that make a big difference in how reliable and compatible your software is.
- I spent some time optimizing Docker images and learning how to speed up program execution with practices like concurrency.
- GSoC also helped me improve my documentation game. Writing weekly progress reports, crafting clear commit messages, and keeping a work log became second nature, and it really pays off in keeping everything organized.
- Attending the weekly community meetings and project calls was a big part of my GSoC experience. They really helped me see the bigger picture and kept me motivated. Plus, these calls were great for making sure everyone was on the same page and moving in the right direction.
Acknowledgements
I want to express my deepest gratitude to everyone who supported me during my GSoC 2024 journey with FOSSology.
First and foremost, I would like to thank my mentors, Shaheem Azmal M MD, Gaurav Mishra, Avinal Kumar and Kaushalendra Pratap whose guidance, patience, and expertise were invaluable. Your encouragement and feedback helped me grow both technically and personally, and I’m incredibly grateful for all the time and effort you invested in my project.
A huge thank you to my family for their unwavering support and understanding throughout this journey. Your belief in me kept me motivated, and I couldn’t have done this without you.
Finally, I’d like to extend my thanks to the entire FOSSology community. From the very beginning, you were welcoming and always ready to help. Working with such a friendly and knowledgeable group made this experience truly rewarding, and I’m proud to have contributed to this amazing project.
Thank you all for making GSoC 2024 such a memorable and transformative experience for me.
Planning for future
- I realize that writing open source code comes with the responsibility to maintain it. And I am more than happy to do so.
- The next major goal for me is to wrap up the dependency scan part of the project; of which NPM dependencies and PHP dependencies are the one’s I am currently working on.
- In the longer run, I plan to keep involved with the community, continue to contribute to open source and most importantly, continue to learn newer things.